Construct, run, and debug workflows from a Jupyter Notebook. Perform simple interactions on the command line. Work across environments and get started quickly with sample workflows using a Python API.
Data management
Data transfer, input selection, and output registration as auxiliary jobs to the workflow.
Error recovery
Retried tasks, workflow checkpoints, remapping, and alternative data sources for staging.
Provenance tracking
Trace a workflow and its outputs, including information about data sources and software used.
Workflows
We have Jupyter based training notebooks available that walk you through creating simple diamond workflow (and more complex ones) using the Pegasus Python API and executing them on ACCESS resources.
-
Why use workflows
Scientific Workflow Management Systems (WMS) such as Pegasus are vital tools for scientists and researchers in a wide range of fields. They provide a structured environment for the design, execution, and monitoring of complex computational tasks, enabling researchers to manage and process large amounts of data efficiently.
Some reasons you should consider using a system like Pegasus WMS:
REPRODUCIBILITY
Document and reproduce your analyses, ensuring their validity, with scientific workflows.
AUTOMATION
Automate repetitive and time-consuming tasks and reduce your workload.
SCALABILITY
Handle large data sets and complex analyses and take on bigger research problems.
REUSABILITY
Build libraries of reusable code and tools that can be adapted by other researchers.See workflow examples
Getting Started with Pegasus
To start you need some Python/Jupyter Notebook knowledge, familiarity with a terminal window, and an ACCESS allocation. How to get an ACCESS Allocation.
-
Get setup
Logon with your ACCESS ID and use Open OnDemand. You specify which allocations you have the first time you logon.
SINGLE SIGN ON WITH YOUR ACCESS ID
All registered users with an active allocation automatically have an ACCESS Pegasus account.
CONFIGURE RESOURCES ONCE
Use Open OnDemand instance at resource providers to install SSH keys and determine location allocation ID.
-
Run workflows on ACCESS
To get started you only need some Python/Jupyter Notebook knowledge, some experience using a terminal window, and an ACCESS allocation.
1
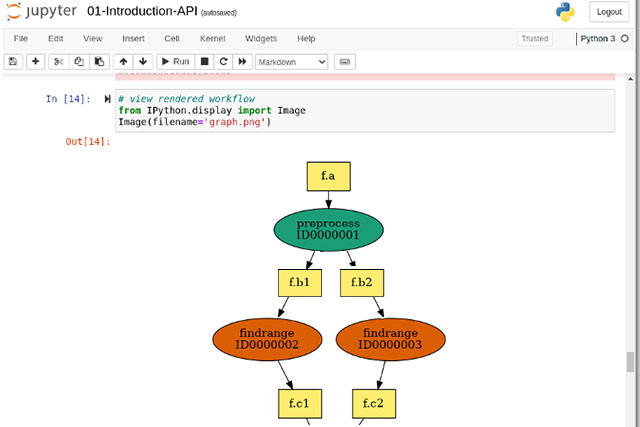
CREATE THE WORKFLOW
- Use Pegasus API in Jupyter Notebook or use our examples
- Submit your workflow for execution,
2
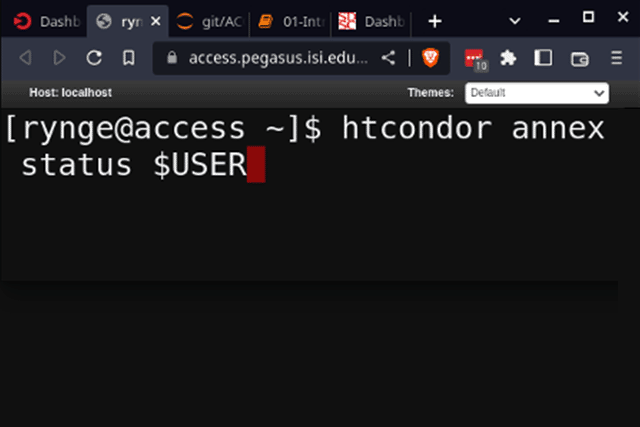
PROVISION COMPUTE RESOURCES
- Use HTCondor Annex tool to provision pilot jobs on your allocated ACCESS resources
3
MONITOR THE EXECUTION
- Follow the workflow execution within the notebook or in the terminal
- You can see what resources you brought in using the terminal