Submission information
Submission Number: 123
Submission ID: 216
Submission UUID: 441ea3e2-ee59-461b-be6c-af1b112aae33
Submission URI: /form/project
Created: Tue, 11/02/2021 - 06:12
Completed: Tue, 11/02/2021 - 06:12
Changed: Mon, 11/21/2022 - 14:26
Remote IP address: 74.103.220.121
Submitted by: Gaurav Khanna
Language: English
Is draft: No
Webform: Project
| Received Sent | 0 |
|---|---|
| Accept and Publish Sent | 0 |
| Project Title | Host-symbiont population genomics - analyzing the intraspecific variability of methanogenic archaeal endosymbionts of genus Methanocorpusculum hosted by a marine anaerobic ciliate Metopus sp. (Metopida) |
| Program | CAREERS |
| Project Image |
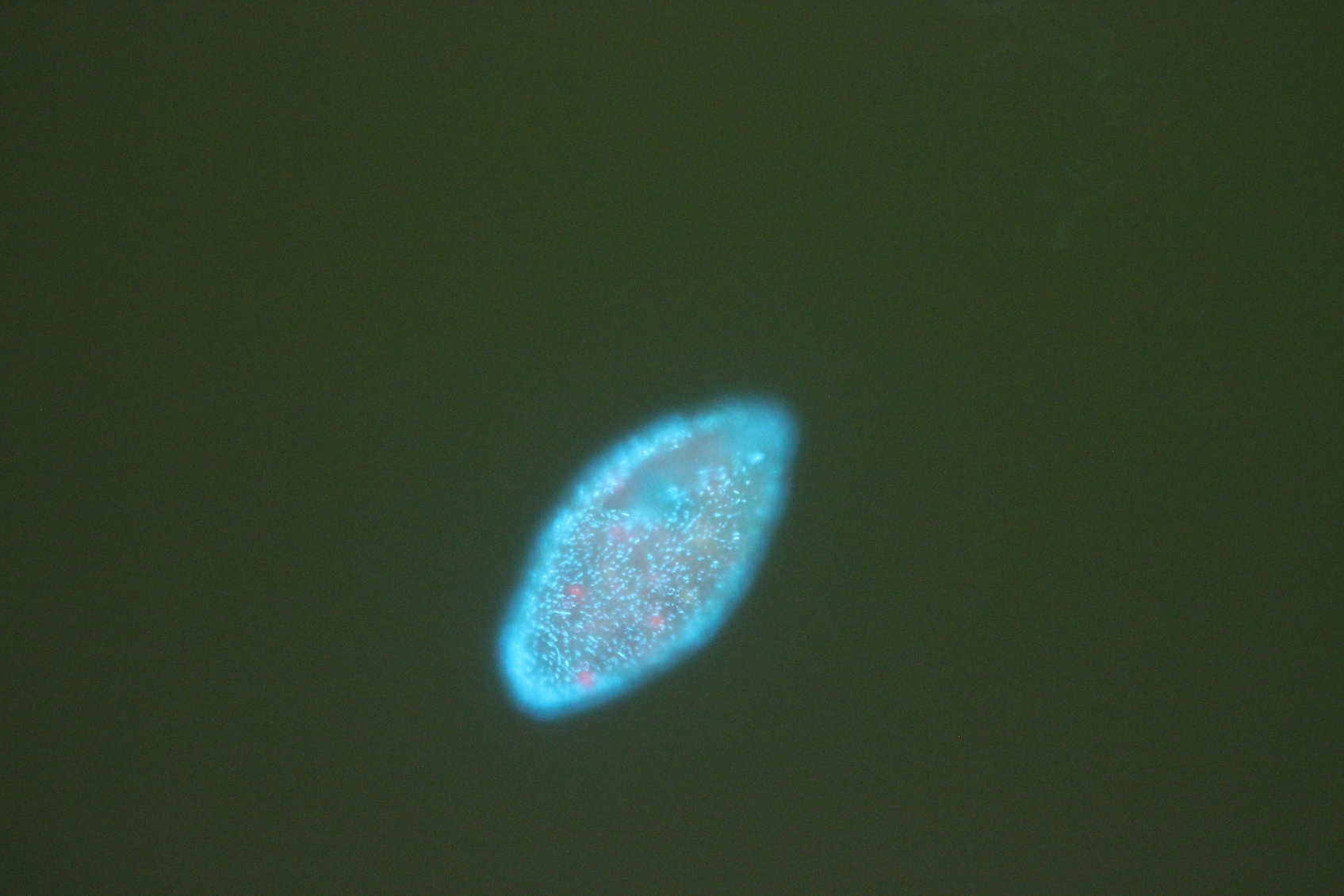
|
| Tags | bioinformatics (277), biology (515), genomics (537) |
| Status | Complete |
| Project Leader | Roxanne Beinart |
| rbeinart@uri.edu | |
| Mentor(s) | Roxanne Beinart |
| Student-facilitator(s) | Johana Rotterova |
| Mentee(s) | |
| Project Description | This project aims to deepen our knowledge of the diversity of archaea-ciliate symbioses that are ubiquitous in anoxic coastal sediments. Using bioinformatic approaches, we plan to assess the intraspecific diversity and population structure of methanogenic archaeal endosymbionts hosted by a marine anaerobic ciliate Metopus sp. (Metopida). We have prepared a nested experimental design for sequencing of 96 single-cell metagenomes of 15 cultivated and uncultivated strains of this ciliate species, isolated from various locations across coastal New England, as well as one from a geographically remote location on the other side of the Atlantic Ocean. Using comparative and population genomics methods, we will uncover host-symbiont specificity patterns, which will provide insight into the evolution of this unique symbiotic relationship. The main goal of this CyberTeams project is to get assistance in setting up these genomic methods on URI and MGHPCC HPC systems for faster throughput. |
| Project Deliverables | The main deliverable of this CyberTeams project is a working and tested workflow to execute genomic methods on URI and MGHPCC HPC systems for faster throughput. |
| Project Deliverables | |
| Student Research Computing Facilitator Profile | |
| Mentee Research Computing Profile | |
| Student Facilitator Programming Skill Level | Some hands-on experience |
| Mentee Programming Skill Level | |
| Project Institution | University of Rhode Island -- Bay Campus |
| Project Address | Rhode Island. 02881 |
| Anchor Institution | CR-University of Rhode Island |
| Preferred Start Date | 01/01/2022 |
| Start as soon as possible. | No |
| Project Urgency | Already behind3Start date is flexible |
| Expected Project Duration (in months) | 6 months |
| Launch Presentation | |
| Launch Presentation Date | 02/09/2022 |
| Wrap Presentation | |
| Wrap Presentation Date | 11/09/2022 |
| Project Milestones |
|
| Github Contributions | |
| Planned Portal Contributions (if any) | |
| Planned Publications (if any) | |
| What will the student learn? | |
| What will the mentee learn? | |
| What will the Cyberteam program learn from this project? | |
| HPC resources needed to complete this project? | |
| Notes | |
| What is the impact on the development of the principal discipline(s) of the project? | Studying symbioses between unicellular eukaryotes (protists) and intracellular methanogenic archaea is critical to our understanding of the evolution of eukaryotic life without oxygen, as well as the impact of these multi-domain holobionts on global methane production. Nearly all anaerobic ciliates, ecologically important protists commonly found in oxygen depleted environments, host methanogenic endosymbionts, but their host-symbiont specificity patterns and intraspecific variability remain largely unknown. We bring the first large-scale genomic analysis of the population structure of both host and symbionts in this partnership, providing key insights into the specificity of these symbioses, and building a foundation for future studies of eukaryotic-prokaryotic liasons in anoxia. |
| What is the impact on other disciplines? | |
| Is there an impact physical resources that form infrastructure? | |
| Is there an impact on the development of human resources for research computing? | |
| Is there an impact on institutional resources that form infrastructure? | |
| Is there an impact on information resources that form infrastructure? | |
| Is there an impact on technology transfer? | |
| Is there an impact on society beyond science and technology? | |
| Lessons Learned | I have learned to work at the URI Andromeda research cluster, how to properly schedule and plan large computing jobs using several nodes but also to back up all data on a regular basis. |
| Overall results | I was able to use and adapt computational software pipelines to assemble and analyze new transcriptomic and large-scale metagenomic data that we have sequenced. I have assembled a eukaryotic transcriptome from the target organism (ciliate hosting archaeal endosymbiont) and genomes from 78 single-cell metagenomic samples from various populations of the archaeal endosymbiont (Methanocorpusculum sp.) from the same ciliate host species. I have analyzed the genomic data to assess the symbiont-host co-diversification and genetic variation. In addition, I have reconstructed mitochondrial genomes from the selected ciliate host populations and performed a mitochondrial phylogenomic analysis to confirm the host phylogeny. |